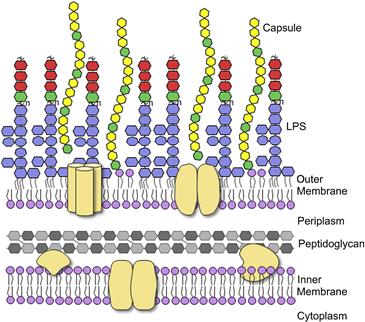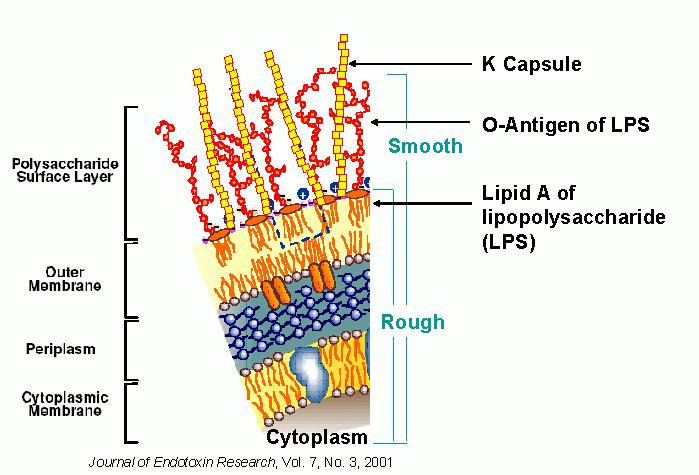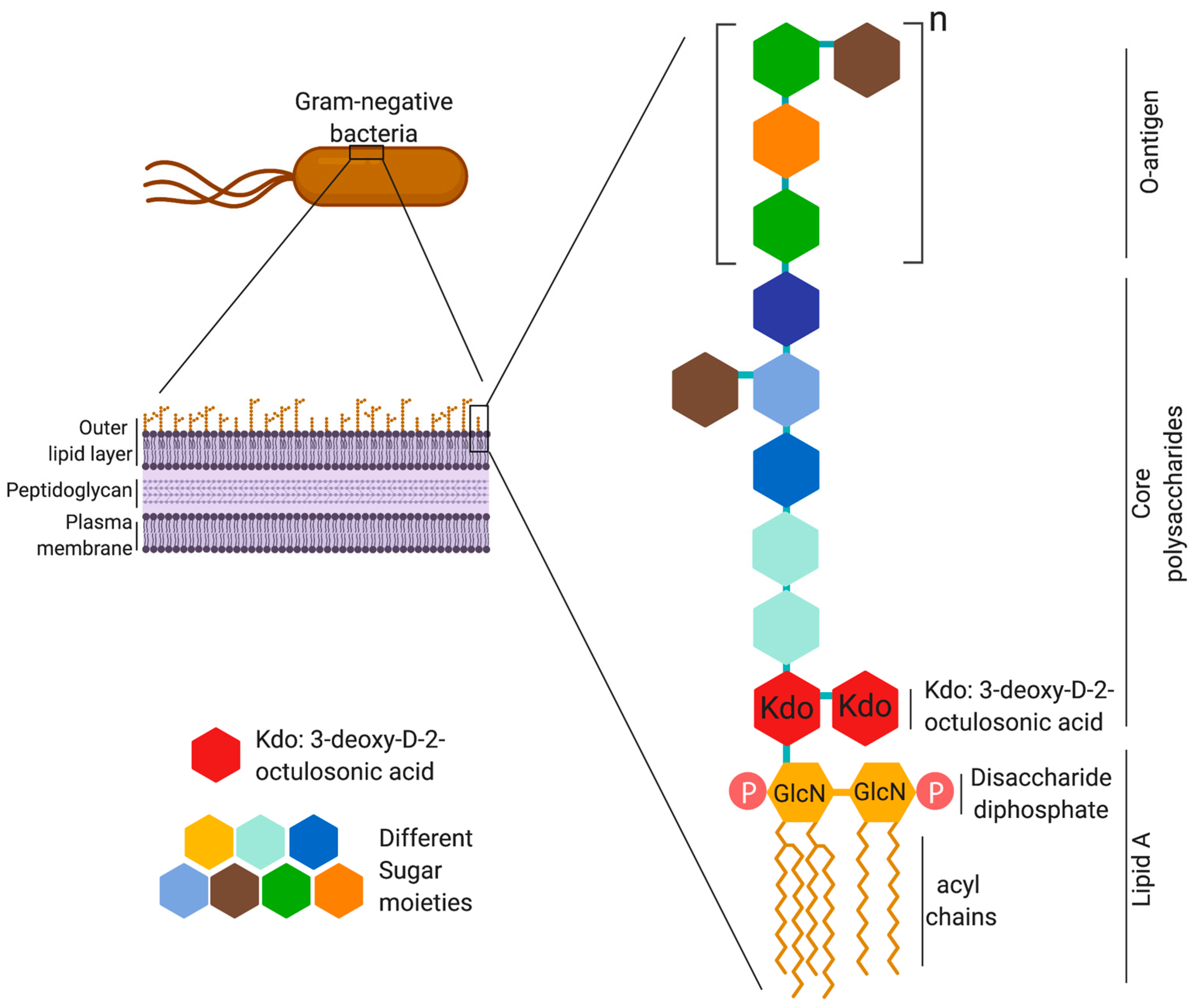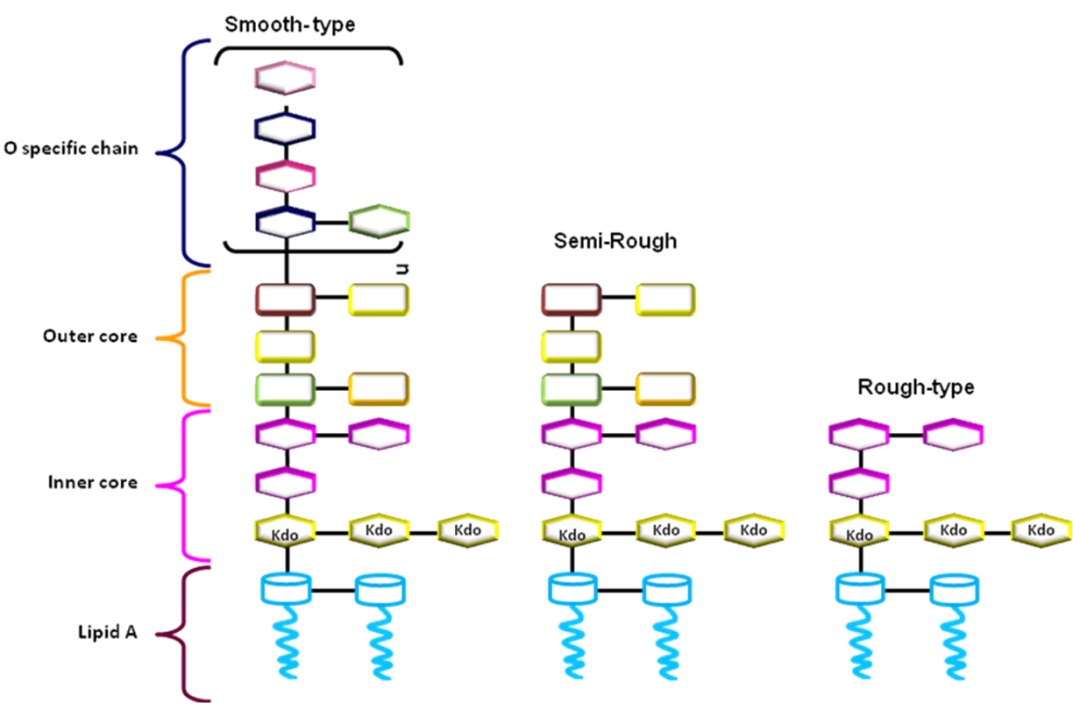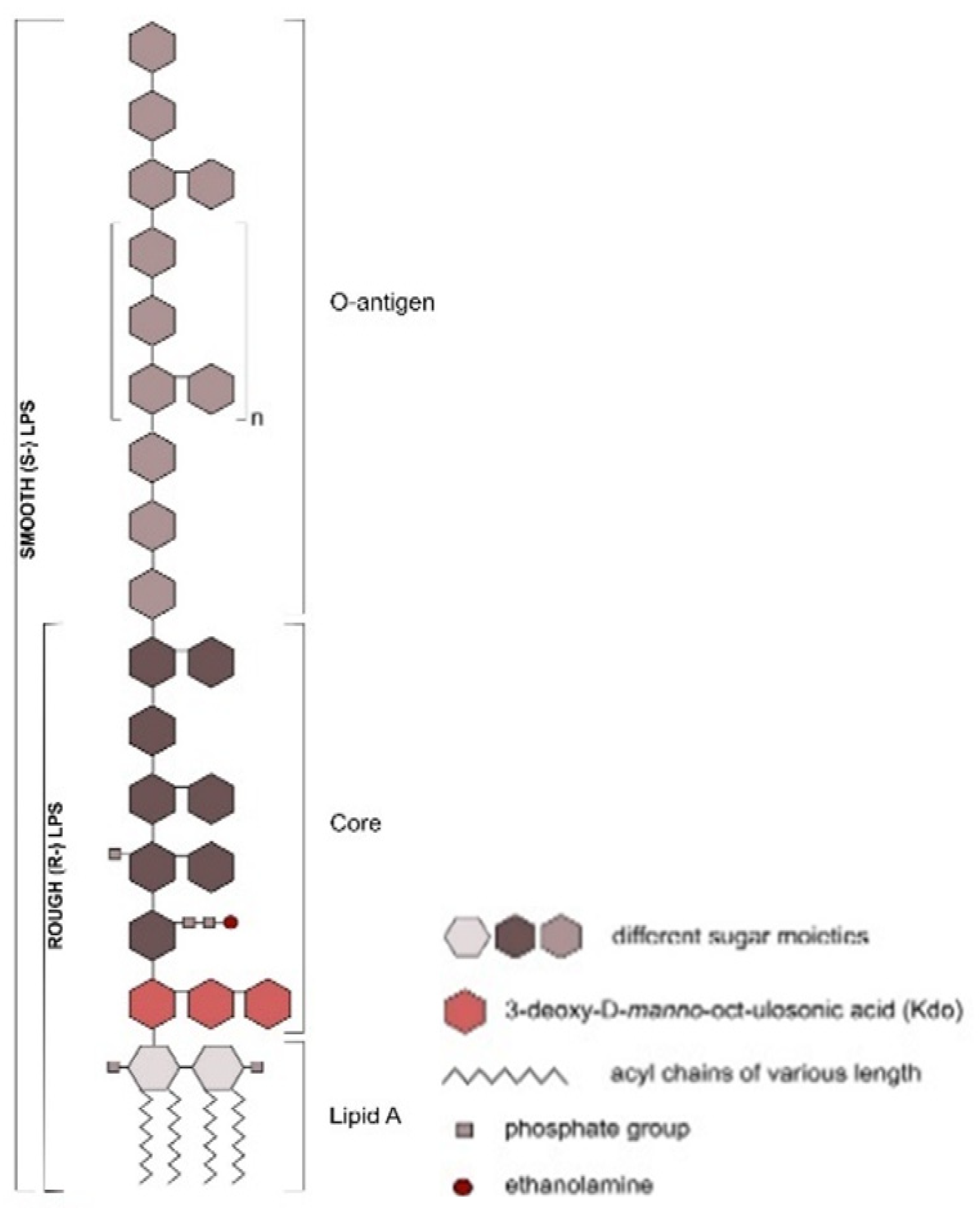
Animals | Free Full-Text | Ruminal Lipopolysaccharides Analysis: Uncharted Waters with Promising Signs | HTML

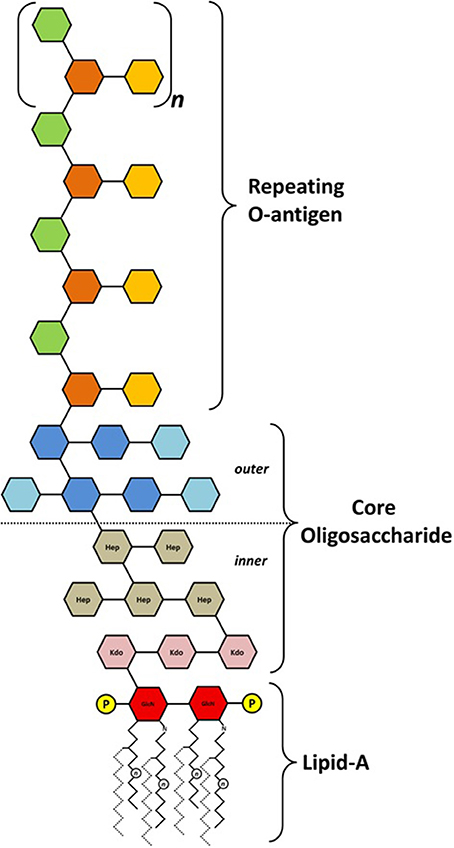
Schematic of the basic structure of lipopolysaccharide. LPS consists of... | Download Scientific Diagram

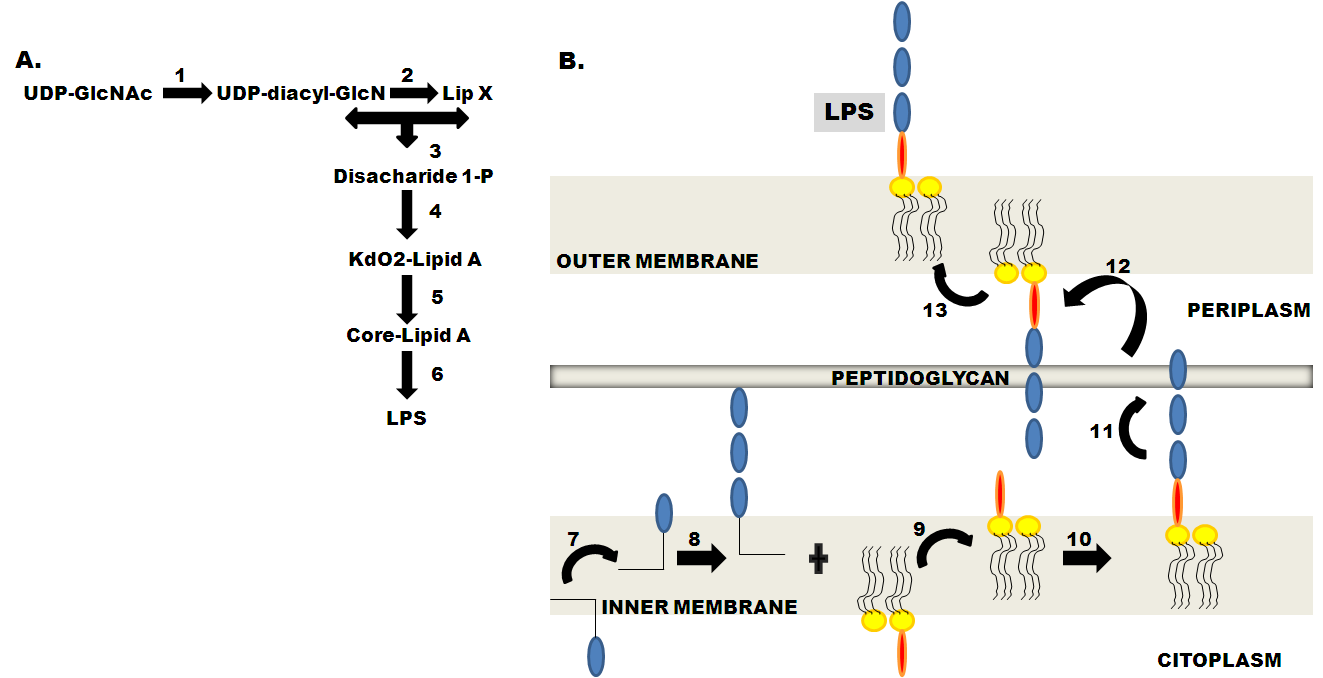
Interrupting Biosynthesis of O Antigen or the Lipopolysaccharide Core Produces Morphological Defects in Escherichia coli by Sequestering Undecaprenyl Phosphate | Journal of Bacteriology
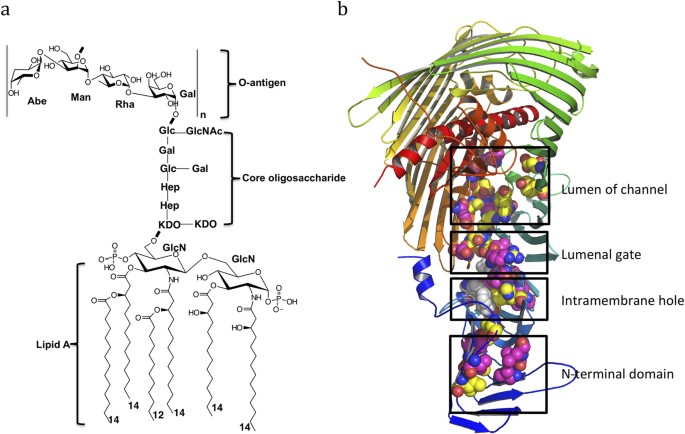
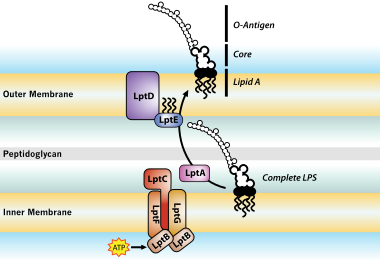
Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane | Scientific Reports

Structural basis for endotoxin neutralization by the eosinophil cationic protein - Pulido - 2016 - The FEBS Journal - Wiley Online Library

Diversity of O-Antigen Repeat Unit Structures Can Account for the Substantial Sequence Variation of Wzx Translocases | Journal of Bacteriology

Lipopolysaccharide O antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a | PNAS
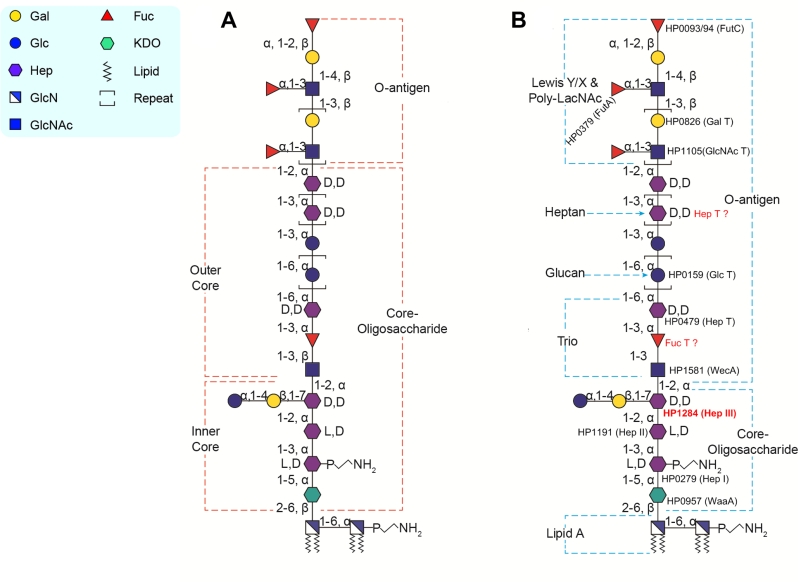
PLOS Pathogens: The redefinition of Helicobacter pylori lipopolysaccharide O -antigen and core-oligosaccharide domains

LPS and O-antigen synthesis. (A) Model representing LPS and O-antigen... | Download Scientific Diagram
PLOS Pathogens: The redefinition of Helicobacter pylori lipopolysaccharide O -antigen and core-oligosaccharide domains
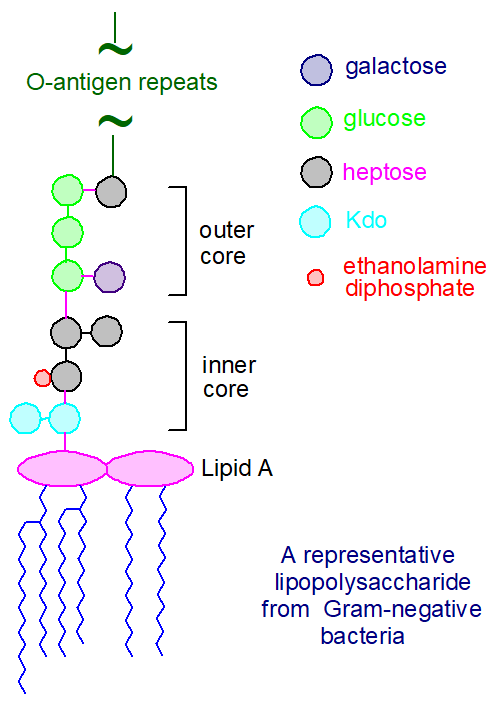
Lipid A and Bacterial Lipopolysaccharides, Gram-negative, endotoxins, lipo-chitooligosaccharides (Nod factors) - structure, occurrence, biochemistry and function

Identification of an O‐antigen chain length regulator, WzzP, in Porphyromonas gingivalis - Shoji - 2013 - MicrobiologyOpen - Wiley Online Library