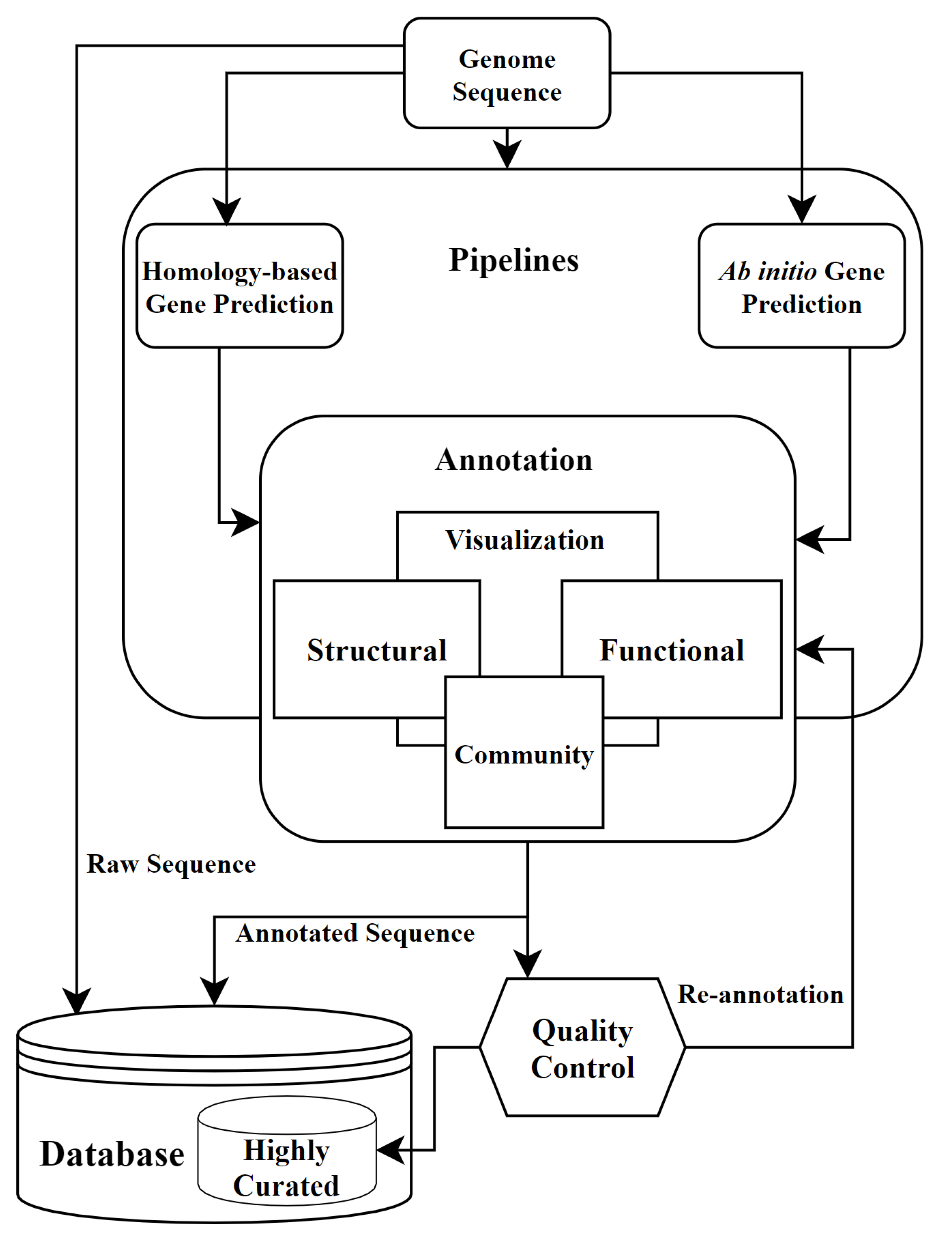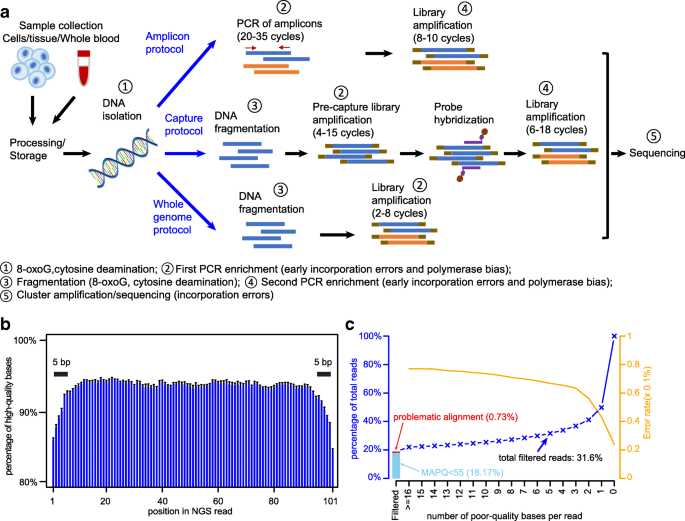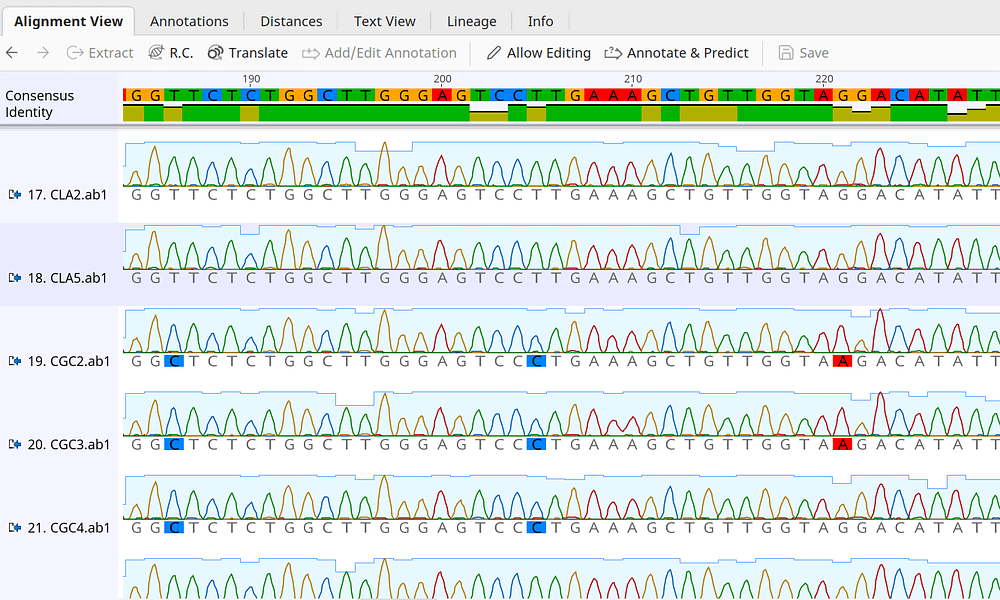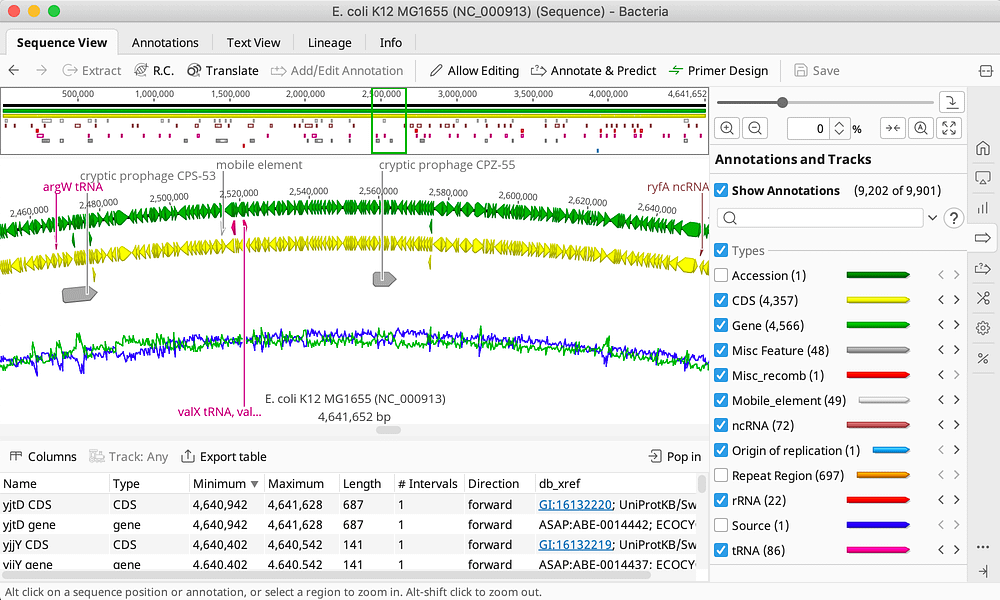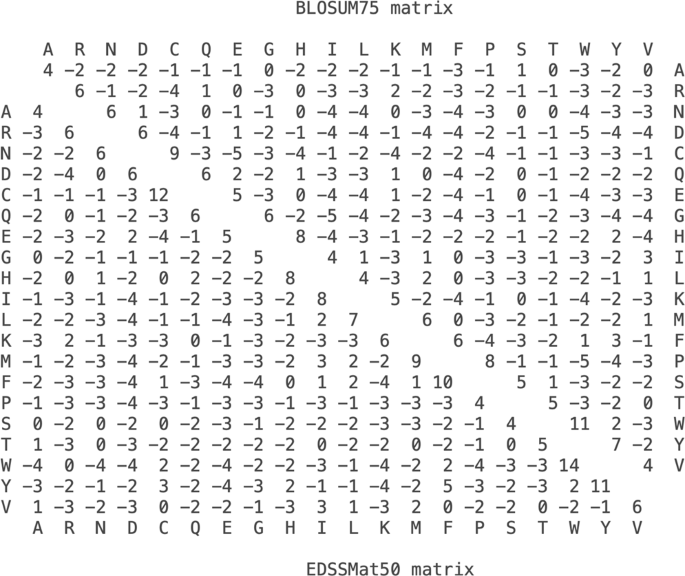
Amino acid substitution scoring matrices specific to intrinsically disordered regions in proteins | Scientific Reports

Tracking the amino acid changes of spike proteins across diverse host species of severe acute respiratory syndrome coronavirus 2 - ScienceDirect

Characteristics of DNA polymerase I from an extreme thermophile, Thermus scotoductus strain K1 - Saghatelyan - 2021 - MicrobiologyOpen - Wiley Online Library

Neural networks to learn protein sequence–function relationships from deep mutational scanning data | PNAS

PepExplorer: A Similarity-driven Tool for Analyzing de Novo Sequencing Results* - Molecular & Cellular Proteomics
Identification and Evolutionary Analysis of Potential Candidate Genes in a Human Eating Disorder - Document - Gale OneFile: Health and Medicine
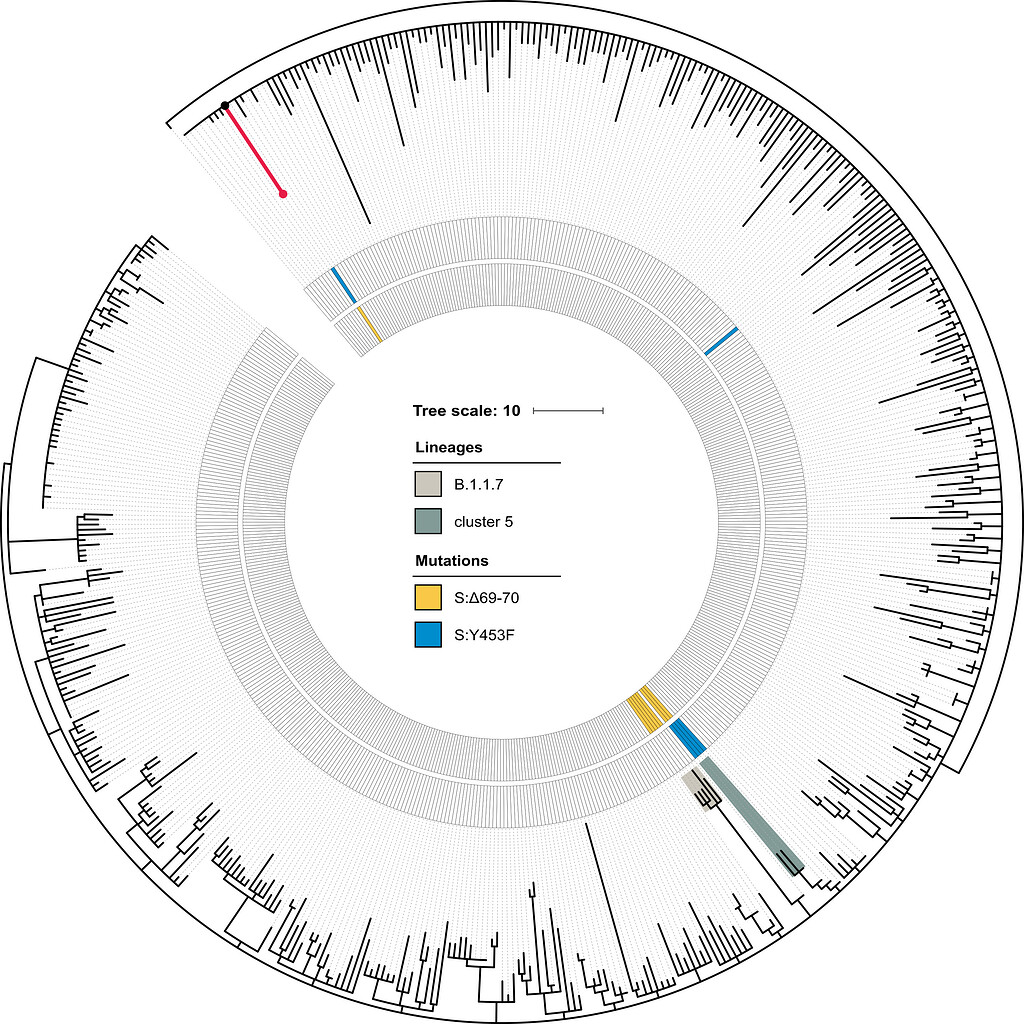
Emergence of Y453F and Δ69-70HV mutations in a lymphoma patient with long-term COVID-19 - nCoV-2019 Genomic Epidemiology - Virological

FastMG: a simple, fast, and accurate maximum likelihood procedure to estimate amino acid replacement rate matrices from large data sets | BMC Bioinformatics | Full Text

How Deep Learning Tools Can Help Protein Engineers Find Good Sequences | The Journal of Physical Chemistry B
rapid generation of mutation data matrices from protein sequences | Bioinformatics | Oxford Academic

Development of Dengue Virus Serotype–Specific NS1 Capture Assays for the Rapid and Highly Sensitive Identification of the Infecting Serotype in Human Sera | The Journal of Immunology