Assessment of ligand binding site predictions in CASP10 - Gallo Cassarino - 2014 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
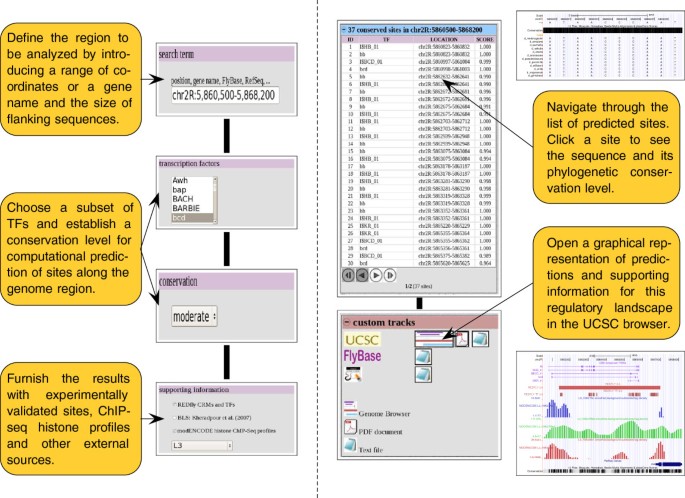
CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes | BMC Genomics | Full Text

Predicted subcellular localization of VC1 by TargetP 2.0 The protein... | Download Scientific Diagram

NetPhos server output for ROP38 phosphorylation sites. (A) The number... | Download Scientific Diagram

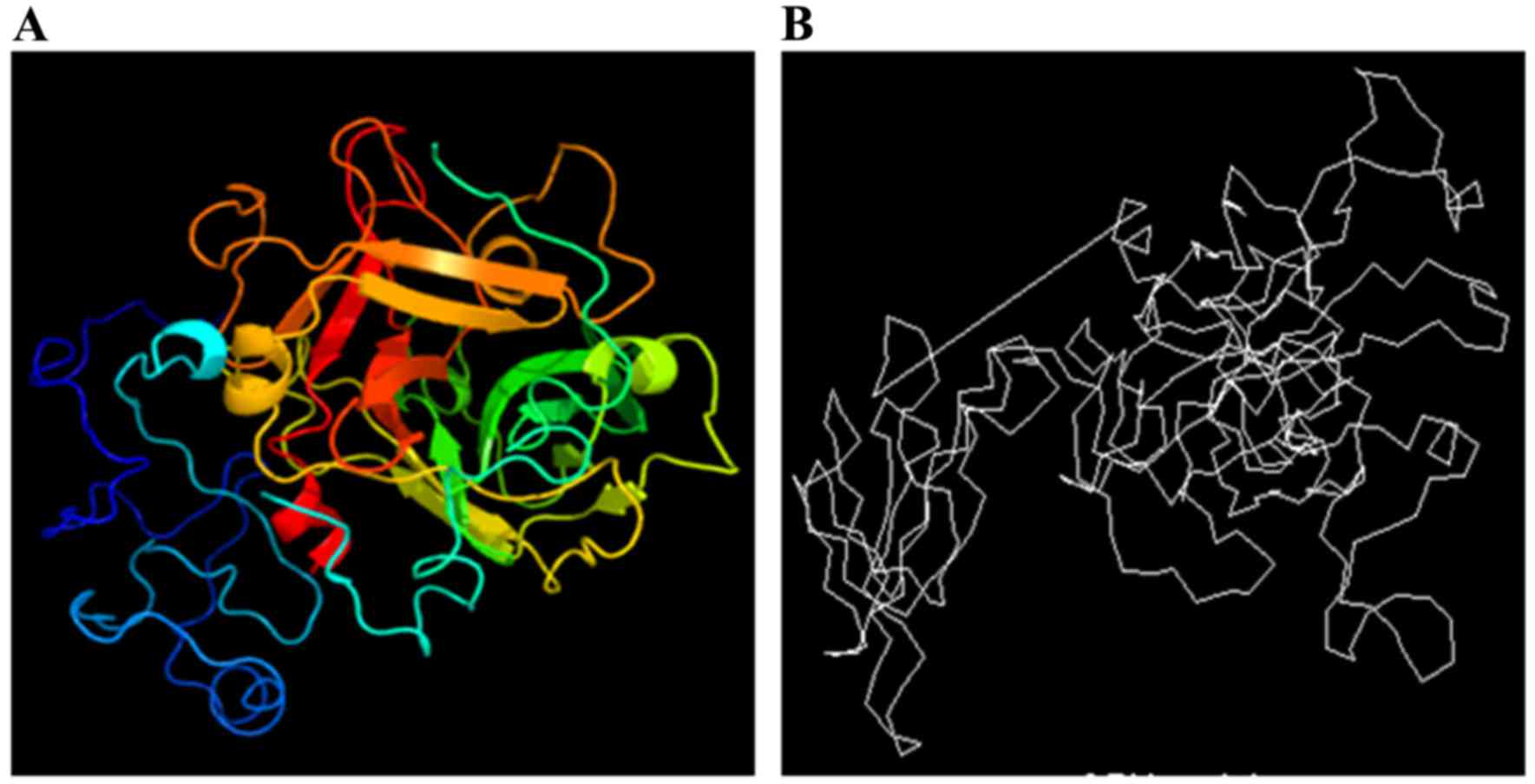
Assessment of ligand binding site predictions in CASP10 - Gallo Cassarino - 2014 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
Center for Biologisk Sekvensanalyse Nikolaj Blom Center for Biological Sequence Analysis BioCentrum-DTU Technical University of Denmark - ppt download
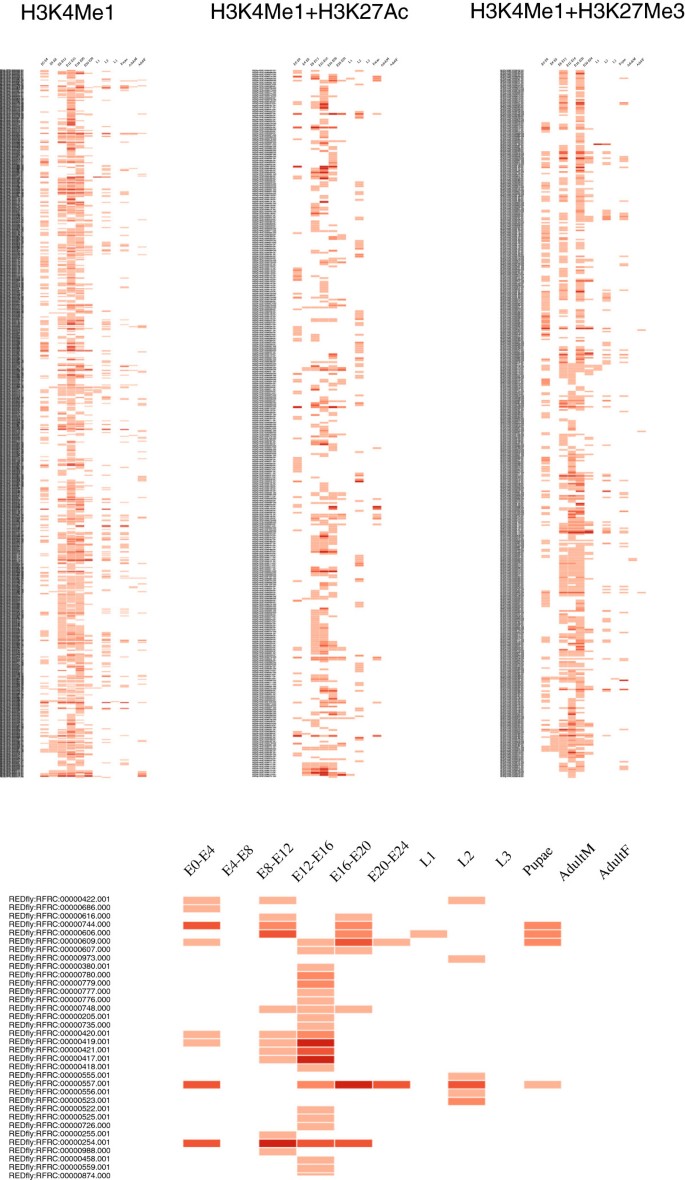
CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes | BMC Genomics | Full Text
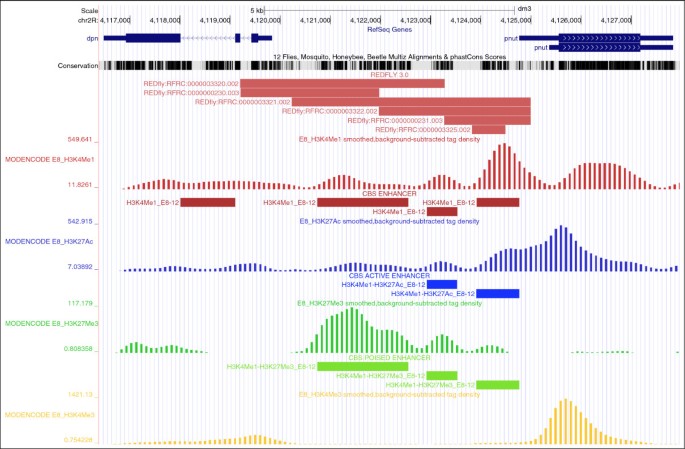
CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes | BMC Genomics | Full Text
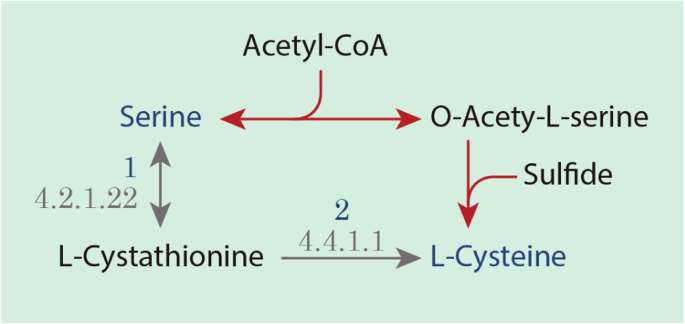
Genome-wide sequencing and metabolic annotation of Pythium irregulare CBS 494.86: understanding Eicosapentaenoic acid production | BMC Biotechnology | Full Text

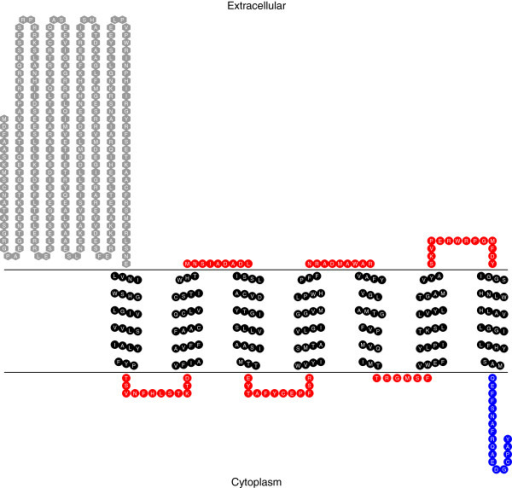
Post-translational modifications of the HYPAI predicted by CBS server.... | Download Scientific Diagram
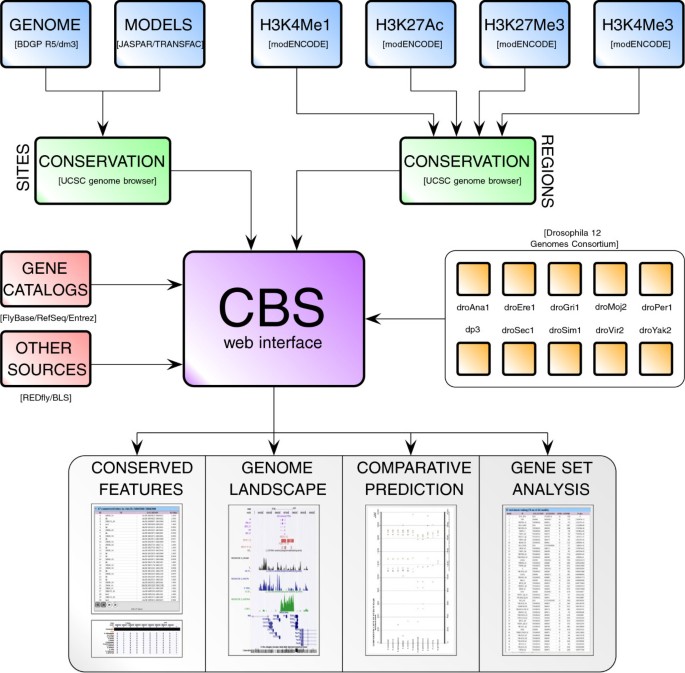
CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes | BMC Genomics | Full Text